GENOMIC MEDICINE, BIOMARKERS AND THE PROGRESS TOWARDS PERSONALISED MEDICINE: THE HEPATITIS C PARADIGM
Michael Carr Ph.D., Clinical Scientist Molecular Reference and Research Unit, National Virus Reference Laboratory, University College Dublin, Belfield, Dublin 4, Ireland
Disclaimer: The views expressed within this article are the author's own and do not necessarily reflect those of the National Virus Reference Laboratory or UCD
ABSTRACT
Hepatitis C virus (HCV) exerts a profound burden of liver disease with global estimates suggesting over 200 million people are infected. Current therapy for chronic HCV infection employs pegylated interferon- (IFN) and the nucleoside analogue ribavirin (RBV). Treatment success is dependent on many factors; including viral genotype, gender, ethnicity, liver histology, baseline viral load and pre- treatment levels of interferon-stimulated genes. Genes influencing treatment response have been identified in cohorts comprised predominantly of the "difficult to treat" HCV genotype 1 and a number of groups independently described genetic variation adjacent to the interleukin 28B gene (IL28B) - encoding interferon- 3 (IFN- 3) which was associated with treatment response and also with spontaneous clearance without treatment. From a clinical point of view, these findings have created great excitement as it has opened the possibility for determination of the IL28B genotype status of patients presenting with chronic HCV infection. Conceivable clinical algorithms have been suggested where individuals with haplotypes associated with HCV spontaneous clearance might be monitored longer whereas patients with haplotypes associated with viral persistence might receive therapy during the acute period and be monitored for a shorter period prior to treatment. Remarkably, new HCV treatment modalities promise a cure with the advent of direct acting antivirals, such as the protease and polymerase inhibitors, despite issues of cost and development of resistance. Nevertheless IL28B genetic testing and prediction of HCV treatment response to IFN/RBV serves to illustrate methodologies which will likely become increasingly common in clinical practice in the future where the treatment is tailored to each patient rather than a one size fits all approach
Article
Introduction
Following the completion of the human genome project in 2003, expectations were raised that this landmark event in human history would herald the rapid translation of new treatments for human genetic disease, a deepening of the understand- ing of the interaction between the human immune system and pathogenic agents and a revolution in personalised medicine where treatments would be tailored to the individual patient [1]. Personalised medicine and the use of biomarkers - definable as quantifiable indicators in the patient of normal biological or pathogenic processes or responses to treatment and/or other therapy - gleaned from advances in basic research offer an alternative to the empirical (trial and error based) treatment approaches.
HEPATITIS C VIRUS
Hepatitis C virus (HCV) was first described in 1989 by Michael Houghton and co-workers [2] and is a hepatotropic member of the family Flaviviridae within the genus Hepacivirus. The Flaviviridae includes the arboviruses (arthropod-borne viruses) dengue and yellow fever virus from which the family derives its name, as the jaundice is characteristic of the latter infection (flavus is Latin for yellow). The viral particle consists of an envelope surrounding a positive polarity RNA genome of some 9600 nucleotides which encodes a polyprotein that is processed by cellular and virally-derived proteases. Staggeringly, 3% of the human population, over 200 million individuals, are estimated to be chronically infected with HCV [3]. This epidemic exerts a profound burden of liver disease and pressure on health care systems particularly as the majority of infections are in the developing world in Africa and Asia. Egypt is a particularly striking example where approximately 14% of the population have serological evidence of previous exposure (anti-HCV antibody) with almost 10% of the population chronically infected (viremic) [4]. This is sadly linked to efforts to eliminate schistosomiasis. HCV positivity strongly correlates with antischistosomal injection treatment before 1986, which unfortunately involved the reuse of contaminated needles. HCV genotype 1 is the pre- dominant genotype in Europe and the United States, so intense research interest has been focused at determining viral and patient-specific markers as well as environmental factors that increase the predictive value of determining the likelihood of treatment success at baseline. One clue was the differences in ethnic responses to treatment where Africans responded less than Europeans who, in turn, had poorer response rates than Asians to standard therapy. This also correlated with rates of clearance in the absence of treatment (so called spontaneous resolvers).
THERAPY FOR HCV INFECTION AND THE SEARCH FOR TREATMENT PREDICTIVE BIOMARKERS
Unlike hepatitis B virus (HBV) no vaccine is currently licensed for HCV. Primary infection with HCV in about 30% of cases results in spontaneous clearance where strong HCV-specific innate and adaptive immune responses control and eliminate the infection. Following establishment of a persistent infection, some 20% of individuals have progressive hepatitis and a further subset progress to worsening liver function with increased fibrosis, cirrhosis and ultimately hepatocellular carcinoma necessitating transplantation.
There are major differences in response rates to standard therapy which currently consists of weight-based pegylated interferon- (PEG-IFN- ) and the nucleoside analogue ribavirin (RBV) for patients chronically infected with HCV. Less than 50% of individuals achieve a sustained virological response (SVR), as determined by the absence of viral RNA six months after treatment cessation with "difficult to treat" HCV genotypes (HCV genotypes 1 and 4) [5]. In contrast, individuals infected with "easy to treat" HCV genotypes 2 and 3 typically achieve SVR rates between 70 and 90% [6]. Hepatologists predicting treatment response and defining treatment duration have relied on clinical and laboratory-derived parameters such as the viral genotype, as described above, the baseline viral load determined by quantitative polymerase chain reaction (qPCR), on treatment viral kinetics, the degree of fibrosis, the extent of hepatosteatosis, the association of female gender with higher clearance rates than males, lack of co-morbidities such as HIV-coinfection, low initial IFN stimulated gene (ISG) expression, and also ethnicity [7].
therapy is associated with significant adverse side effects, is prolonged in duration and expensive
IFN- has been used for more than two decades for treatment of HCV infection, however, therapy is associated with significant adverse side effects, is prolonged in duration and expensive. This has stimulated the search for biomarkers that could potentially be employed to predict an individual’s likelihood of achieving a successful treatment outcome prior to commencement of therapy. In order to explain the recent ground-breaking advances in prospects for personalised therapy for HCV-infected patients we must first discuss the technology and how it has lead to the development of biomarkers to predict treatment response that ultimately improve patient care.
GENOME WIDE ASSOCIATION STUDIES
The complete DNA sequence of a genome from person to person is thought, by various estimates, to differ by approximately 0.1%, which accounts for some 3 million genetic differences within the 3 billion base pairs of the haploid human genome. This inter-individual variation is largely thought to be of a neutral nature whereby the vast majority of this genetic variation does not alter amino acids within proteins and has no discernible phenotypic effect. However, a smaller subset of this variation between individuals can occur within genes leading to changes in the encoded protein (non-synonymous mutations) within regulatory regions, such as promoters, that influence gene expression or splicing or other post-transcriptional effects. When this variation is greater than 1% within a human population the site is termed a single nucleotide polymorphism (SNP).
Rather than studying genetic variation in a single gene derived from a list of plausible gene candidates between cases and control groups, or a small subset of potential target markers, there are alternative strategies which analyse susceptibility loci without a priori hypotheses which by virtue of their essentially random nature offer the potential to uncover new pathways into the aetiogenetic pathogenesis of disease. Genome wide association studies (GWAS) are one such unbiased approach, which analyse gene frequencies of SNP’s across the genome in individuals with the disease or trait of interest by statistical comparison with a suitable control group. GWAS’s have been increasingly used in medical research since the sequencing of the human genome to analyse complex, polygenic groups of human disease such as rheumatoid arthritis, atherosclerosis, Crohn's disease and type 2 diabetes mellitus. The general theme of the results from these diverse areas of study is a complex interaction of small effect genetic susceptibility loci (host genotype) with, of course, environmental factors that lead to disease (host phenotype).
INTERLEUKIN 28B (IL28B) AND HCV CONTROL
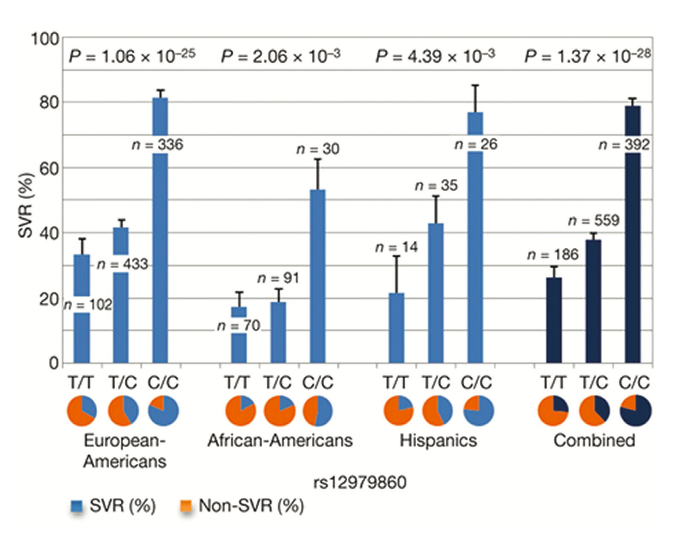
Figure 1. Genetic variation in IL28B predicts HCV treatment-induced viral clearance. Data are presented for each group as percentages + standard error of the mean. Reprinted from Nature 2009, 461:399-401, copyright 2009 with permission from MacMillan Publishers Ltd [8]
In this regard, a notable exception was the strong and unambiguous genetic association uncovered by four independent groups in 2009, conducting research into host genetic factors influencing treatment response rates in chronically HCV-infected patients [8-12]. Their cohorts were comprised predominantly of the "difficult to treat" HCV genotype 1 and strikingly, each study described SNP’s adjacent to the interleukin 28B gene (IL28B) -encoding interferon- 3 (IFN- 3) on the long arm of human chromosome 19 (19q13.13). This was associated with clearance following standard therapy and also, importantly, with spontaneous clearance without treatment. In this landmark study, Ge and colleagues [8] compared the allele frequencies of 600,000 SNPs by GWAS from 1,137 individuals of differ- ing ethnic backgrounds (African, Hispanic and European) with persistent HCV who were treated for 48 weeks with standard therapy and then followed for 24 weeks after discontinuation of treatment. A remarkably strong genetic influence was found for individuals homozygous (CC) for the rs12979860 SNP and treatment-induced clearance. No other SNP’s outside the IFN-cluster approached genome-wide significance for predicting treatment response. Crucially this protective effect of the C allele was seen across the three different ethnic groups and when combined data for the three groups was analysed together [Figure 1] individuals carrying the CC genotype had a sustained virological response (SVR) almost three times higher than the minor homozygote TT poor response genotype (78%/28%). Furthermore, the effect of the T risk allele appeared to be dominant as heterozygosity (CT) lead to a >2-fold decrease (38% SVR rate) in the likelihood of achieving a post- treatment SVR compared to CC homozygotes. The authors compared the known baseline predictors for patient treatment response with the magnitude of this newly uncovered effect for IL28B genotype and concluded that genotyping of this SNP is associated with the most substantial influence on treatment response.
This seminal work was quickly followed by three other studies and the association of genetic variation near IL28B on predict- ing treatment response rates was further strengthened. Tanaka and colleagues [9] analysed by GWAS the genetic influence of 142 Japanese individuals infected with HCV-1, comprising of 78 treatment non-responders and 64 who achieved an SVR; and identified a risk allele (GG) for the SNP rs8099917 8.9kb upstream of the IL28B gene strongly associated with null virological response. Specifically, homozygous carriers of rs8099917 G allele had >2 fold more likelihood to fail to achieve an SVR compared with heterozygotes and the TT major homozygote responder genotype. Suppiah and co-workers [10] in a larger study also identified rs8099917 as the statistically most significant SNP influencing treatment response in 293 Australians of European ancestry, made up of 162 non- responders and 131 with SVR. They validated their findings with 555 patients from Germany, Italy, Australia and the UK. Finally, the Swiss HCV and HIV cohort study [11] analysed 465 individuals including individuals with HCV genotypes 1-4 and again identified rs8099917 as a predictor of treatment response, with the strongest effects in HCV genotypes 1 and 4 infected patients. The lowest carriage rate of the rs8099917 risk allele GG (24%) was in individuals with spontaneous clearance, in 32% of chronically infected who responded to treatment and in 58% who did not clear the virus on treatment. An important technical point to note is that that the three studies above describing the association of rs8099917 with SVR did not contain or had limited representation of the rs12979860 SNP unearthed by Ge and co-workers but the latter SNP has since been replicated independently by a number of other centres and these markers are in strong linkage disequilibrium (i.e. the genotypes at each locus are not independent of one another).
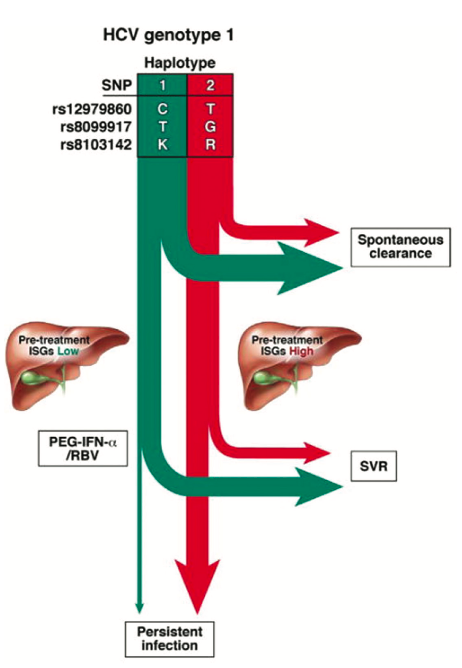
Figure 2. Predicted natural course of HCV infection in the absence of therapy and treatment response based on IL28B haplotypes. Employing IL28B SNPs alone or together allows allele and haplotype tagging associated with a greater likelihood of HCV treatment response spontaneous clearance (depicted by the green arrows) or viral persistence (in red). Reprinted from Gastroenterology 2010, 139:1865-76, copyright 2010 with permission from Elsevier [17]
As many of the SNPs identified by the GWAS studies are located upstream of the IL28B gene - rs12979860 for example lies 3kb upstream of the gene start within a CpG island promoter - this immediately suggested that individuals with the poor response alleles may have lower levels of expression of interferon- 3 (since the IL28B gene encodes for interferon- 3). This has indeed been demonstrated by two groups (looking at the rs8099917 SNP) in peripheral blood mononuclear cells (9,10). This suggests that an attenuated antiviral response to HCV infection could lead to the establishment of a persistent infection, however, Ge and co-workers found no significant association between the rs12979860 SNP and IL28B expression [8]. Interestingly, baseline levels of the downstream mediators of the interferons, the ISGs (interferon stimulated genes), have been inversely correlated with treatment success. That is to say that, paradoxically, individuals with higher ISG expression pre-treatment have poorer treatment success rates so the genetic effects of the risk alleles could be associated with higher ISG expression [13,14]. Even more bizarrely, the good treatment response markers also correlate with higher pre- treatment HCV RNA levels [8]. However, based on a previous study showing that ISG expression may modulate the response to PEG-IFN- [15], this is potentially explicable by patients with higher baseline viral loads having lower basal levels of hepatic ISG expression. These patients with low pre-treatment ISG levels when stimulated with standard therapy then show greater upregulation of ISGs from basal levels and hence better treatment response. In contrast, patients with higher basal levels of hepatic ISG expression gained no benefit from treatment with PEG-IFN- [8].
Clearly all the GWAS studies point to IFN- as central to the control of HCV infection [16]. There are considerable similarities between the modes of action of IFN- and the IFN-’s and the downstream signalling pathways in particular [17, Figure 2]. However, while IFN-receptors which are broadly expressed on most cell types, including leukocytes, IFN- receptors are largely restricted to cells of epithelial origin [18]. Furthermore, the IFN-’s have been shown to inhibit the replication of HCV, HBV and the influenza A virus in vitro [19,20,21] and this has stimulated interest in the IFN-'s as potential therapies in the infectious disease and cancer settings [22].
CLINICAL VIEWPOINT
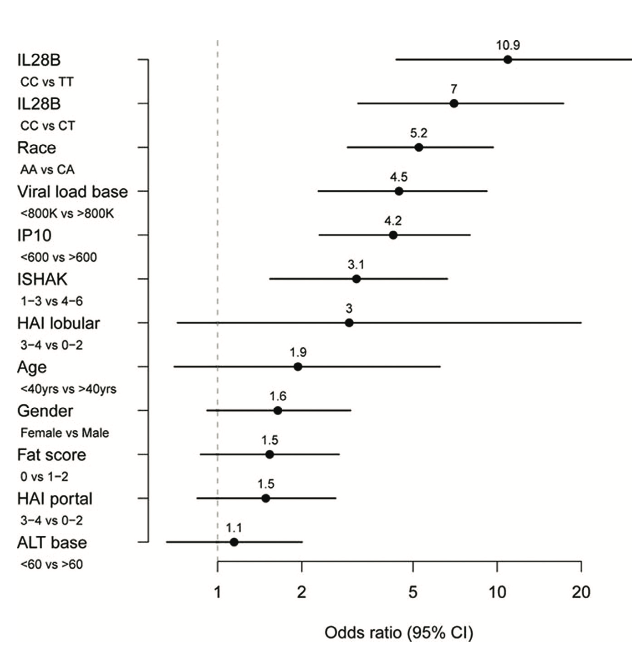
Figure 3. Predictors of sustained virological response to PEG-IFN- and ribavirin therapy.
Odds ratios were calculated from a logistic regression model including IL28B genotype and baseline (pretreatment) measurements of IP-10, HCV viral load, fibrosis stage (ISHAK), age, gender, alanine transaminase (ATL), steatosis (fat score), and portal and lobular histologic activity index (HAI). Reprinted from Hepatology, 2011, 53:14-22, copyright 2011 with permission from John Wiley and Sons Inc. [23]
From a clinical point of view, these basic research findings have created great excitement among clinicians and scientists alike as it has opened the possibility for determination of the IL28B genotype status of patients presenting with chronic HCV infection. Determination of IL28B genotype in replication studies has shown this locus to be the highest independent predictor of treatment response eclipsing viral genotype, pre-treatment (baseline) viral load, liver histology and ethnicity [23, Figure 3]. Conceivable clinical algorithms have been suggested where individuals with a haplotypes associated with HCV clearance in the absence of therapy might be monitored longer, since they are more likely to spontaneously clear the virus whereas patients with haplotypes associated with viral persistence might receive therapy during the acute period and be monitored for a shorter period prior to treatment [17]. The association between kinetics of HCV response to IFN treatment and IL28B genotype might also be used to identify patients that require shorter durations of therapy. The finding that individuals with poor response alleles have lower expression of IFN- suggests that a direct protein based replacement therapy of IFN- in individuals with the poor response alleles may be advantageous. The highly related cytokine, interleukin 29 (IL29, interferon- 1) is in clinical trials [16]. This drug has demonstrably lower toxicity than the current IFN- based regime and importantly exhibited robust antiviral effects [24]. This is likely due to the expression pattern of IFN- receptors, which are predominantly restricted to cells of epithelial origin such as hepatocytes, keratinocytes and bronchial epithelial cells [25]. The decreased side effects, such as reduced bone marrow destruction and flu-like effects common with current standard therapy, is likely attributable to the lower levels of expression of type III (IFN- ) receptors in the bone marrow and in the brain [25].
The recent licensure of HCV protease inhibitors (telaprevir and boceprevir) and other direct acting antivirals (DAAs) in the pipeline (such as further protease and polymerase inhibitors) may render IL28B testing obsolete as they operate independently of IL28B haplotype. However, as with HIV-infected individuals on antiretroviral therapy, the emergence of resistant mutants will need to be evaluated [26]. In the short term, due to the high initial cost of the currently licensed DAAs, IL28B testing may be utilised to identify individuals carrying alleles predicting poor response to standard therapy for treatment with the new agents, whereas the favourable response allele carriers may obtain existing PEG-IFN- /RBV. Furthermore, the knowledge that this form of genetic test highly predicts treatment response can help patients to be encouraged to commence therapy and reassure them during a long treatment course with side effects. A new polymerase inhibitor (PSI-7977) has shown 100% virologic response rates when taken with RBV or three separate PEG-IFN- regimes [27] suggesting a cure for HCV could be on the near horizon only shortly over two decades since the agent’s original identification, which is a remarkable achievement for the medical and scientific communities. The likelihood is that PEG-IFN- /RBV-free DAA-only regimes will go through Phase III clinical trials in 2012.
“What is clear is that personalised genomics will become increasingly common in the future in many areas of medicine. ”
What is clear is that personalised genomics will become increasingly common in the future in many areas of medicine. Whole genome DNA sequencing technology is still prohibitively expensive and technically demanding for the widespread application in the clinical setting. This is expected to change rapidly, however, and the benchmark of the ‘$1,000 genome’ may well have already been reached (28). This is almost beyond com- prehension when you consider that the entire Human Genome Project started in 1990, was finalised at 99.99% coverage in 2003 and cost $3 billion. The advent of genomic medicine is thus fast approaching and its direct impact on patient care is al- ready here, where diagnostics and therapy-related diagnostics (so called theranostics) will be revolutionised by the advances in basic science.
Thomas S. Kuhn argued in The Structure of Scientific Revolutions (1962) that the accepted model of scientific advance involving a slow, linear, incremental accumulation of knowledge was inconsistent with the facts and an episodic, distinctly non- linear model of scientific innovation where revolutions occurred within disciplines was more reflective of the actual workings of the scientific enterprise (29). Kuhn coined the term paradigm shift to denote this model and there appears some similarities to what we are seeing now with the advent of genomic and more personalised medicine. The translation of the vast amounts of genetic information obtained from genomics into medical practice and improving patient care is the ultimate rationale under- pinning the human genome project and will become increasingly more common in clinical practice in the future.
References
1. Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010; 363:301-4.
2. Houghton M. The long and winding road leading to the identification of the hepatitis C virus. J Hepatol. 2009; 51:939-48.
3. Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005; 5:558-67.
4. Nguyen MH, Keeffe EB.Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin Gastroenterol Hepatol. 2005; 3:S97-S101.
5. Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL Jr, Häussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002; 347:975-82.
6. Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiff- man M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001; 358:958-65.
7. Diagnosis, management, and treatment of hepatitis C: an update. Ghany MG, Strader DB, Thomas DL, Seeff LB; American Association for the Study of Liver Diseases. Hepatology. 2009; 49:1335-74.
8. Ge et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009, 461:399-401.
9. Tanaka et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 2009, 41:1105-1109.
10. Suppiah et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 2009, 41:1100-1104.
11. Rauch et al. Genetic variation in IL28B Is Associated with Chronic Hepatitis C and Treatment Failure - A Genome-Wide Association Study. Gastroenterology 2010, 138:1338-1345.
12. Thomas et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009; 461:798-801.
13. Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005; 128:1437-44.
14. Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, Schweigler LM, Theodore D, Zacks SL, Liang TJ, Fried MW. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007; 46:1548-63.
15. Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008; 105:7034-9
16. Kelly C, Klenerman P, Barnes E. Interferon lambdas: the next cytokine storm. Gut. 2011; 60:1284-93
17. Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010; 139:1865- 76.
18. Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN- ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008; 4:e1000017.
19. Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon- and interferon-stimulated genes in primary liver cultures. Hepatology. 2011; 54:1913-23.
20. Hong SH, Cho O, Kim K, Shin HJ, Kotenko SV, Park S. Ef- fect of interferon-lambda on replication of hepatitis B virus in human hepatoma cells. Virus Res. 2007; 126:245-9.
21. Jewell NA, Cline T, Mertz SE, Smirnov SV, Flaño E, Schin- dler C, Grieves JL, Durbin RK, Kotenko SV, Durbin JE. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol. 2010; 84:11515-22.
22. Lasfar A, Abushahba W, Balan M, Cohen-Solal KA. Interferon lambda: a new sword in cancer immunotherapy. Clin Dev Immunol. 2011; 2011:349575.
23. Darling JM, Aerssens J, Fanning G, McHutchison JG, Gold- stein DB, Thompson AJ, Shianna KV, Afdhal NH, Hudson ML, Howell CD, Talloen W, Bollekens J, De Wit M, Scholliers A, Fried MW. Quantitation of pretreatment serum interferon-inducible protein-10 improves the predictive value of an IL28B gene polymorphism for hepatitis C treatment response. Hepatology. 2011; 53:14-22.
24. Ramos EL. Preclinical and clinical development of pegylated interferon-lambda 1 in chronic hepatitis C. J Interferon Cytokine Res. 2010; 30:591-5.
25. Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res. 2010; 30:555-64.
26. Clark PJ, Thompson AJ, McHutchison JG. IL28B genomic- based treatment paradigms for patients with chronic hepatitis C infection: the future of personalized HCV therapies. Am J Gastroenterol. 2011; 106:38-45.
27. Bourlière M, Khaloun A, Wartelle-Bladou C, Oules V, Portal I, Benali S, Adhoute X, Castellani P. Chronic hepatitis C: Treatments of the future. Clin Res Hepatol Gastroenterol. 2011; 35 Suppl 2:S84-95.
28. http://medicalxpress.com/news/2012-01-davos-wowed- device-code-life.html
29. Kuhn TS: The Structure of Scientific Revolutions. 3rd edition. Chicago and London: University of Chicago Press; 1996.